patcHwork is free and open to all users and there is no login requirement.
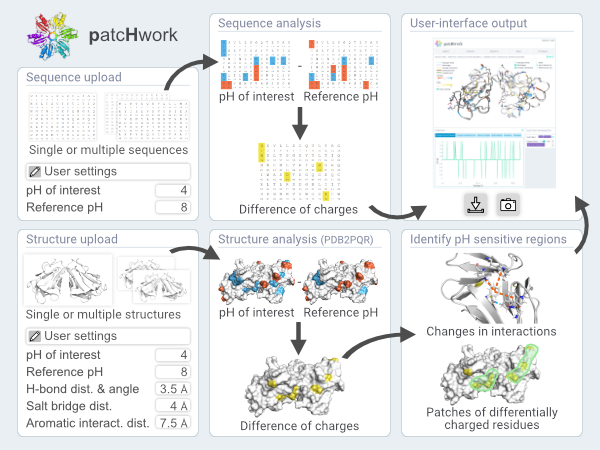
To gain a deeper mechanistic understanding of the effect of pH changes on a protein of interest, patcHwork performs an analysis on submitted 3D protein structures using the PDB2PQR and PROPKA tools. The graphic user interface of patcHwork makes it possible to easily locate pH-sensitive residues on the protein structure at two defined pH values as well as to visualize salt bridges, hydrogen bonds and aromatic interactions that change upon pH shift. Furthermore, patcHwork offers the possibility to identify patches of pH-sensitive residues that are in close proximity on the protein structure, as they are likely to have a major impact on protein activity.
patcHwork: a user-friendly pH sensitivity analysis web server for protein sequences and structures Mirko Schmitz, Anne Schultze, Raimonds Vanags, Karsten Voigt, Barbara Di Ventura, Mehmet Ali Öztürk Nucleic Acids Research 2022 50 (W1), W560-W567
References:
PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations
Todd J. Dolinsky, Paul Czodrowski, Hui Li, Jens E. Nielsen, Jan H. Jensen, Gerhard Klebe, and Nathan A. Baker's Nucleic Acids Research 2007 35 (Web Server issue), W522-5.
PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions
Mats H. M. Olsson, Chresten R. Søndergaard, Michal Rostkowski, and Jan H. Jensen Journal of Chemical Theory and Computation 2011 7 (2), 525-537
NGL viewer: web-based molecular graphics for large complexes
Alexander S Rose, Anthony R Bradley, Yana Valasatava, Jose M Duarte, Andreas Prlić, and Peter W Rose Bioinformatics 2018, 34 (21), 3755-3758
Derivation and use of a formula to calculate the net charge of acid-base compounds. Its application to amino acids, proteins and nucleotides
José C. Cameselle, João Meireles Ribeiro, and Antonio Sillero Biochemical Education 1986, 14 (3), 131-136




